If you prefer videos, there's a version of the tutorial here : YouTube GCode_comments tutorial.
Introduction :
If you're into 3D printing, then you may have been in the situation that you want to make another copy of a print you made months ago, and you don't want to re-slice the model, because you made lots of fine adjustments in the slicer that you have forgotten, but you also don't want to just throw the G-code into the printer because you're not sure what nozzle and material it was sliced for.
One solution to this is to manually construct file names that include all this information, but that is annoying to do every time. There is a plug-in for Cura that will do that construction for you automatically (Gcode Filename), but the result is still rather clumsy, and besides it doesn't work under my OS.
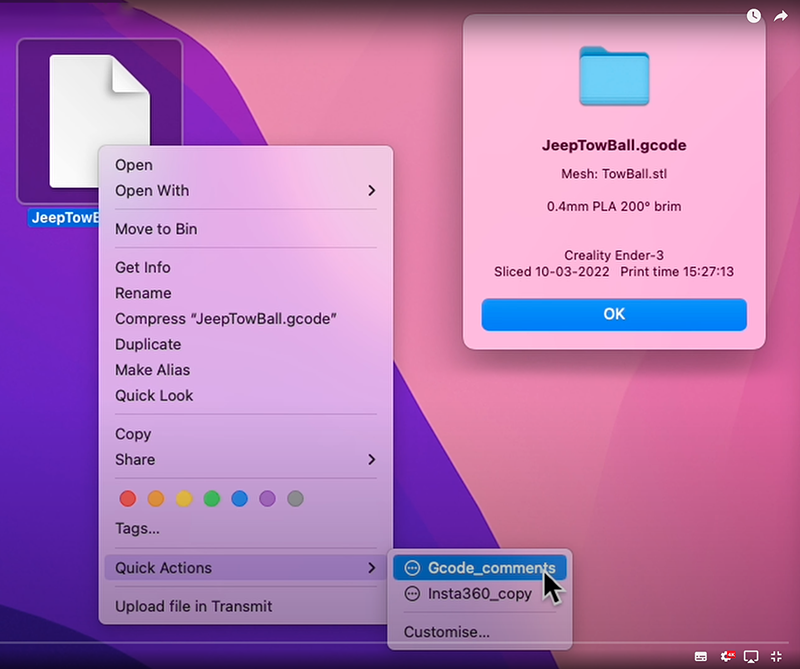
So the solution I use is to get Cura to write the most interesting slicer conditions into the G-code file as comments, and then use Apple's Automator function to extract those comments and show them in an Alert pop-up when I right-click a file and choose the function 'Gcode_comments".
Writing the comments :
In Cura you can add comments to your G-code by going to : Settings>Printer>ManagePrinters>MachineSettings
and then in the "End G-code" window add something like the following :
;--------------------------------------------------
; >>>Machine name: {machine_name}
; >>>Nozzle: {machine_nozzle_size}
; >>>Material: {material_name}
; >>>Speed: {speed_print}
; >>>Temperature: {material_print_temperature}
; >>>Adhesion: {adhesion_type}
; >>>Print time: {print_time}
; >>>Slice date: {date}
;--------------------------------------------------
The semicolon denotes a comment, the chevrons are a string that I have chosen to uniquely identify my comments, and the names in braces are variables that Cura knows to replace with actual values.
Extracting the comments with Automator :
Open the Automator app in OS X. Select File>New, Choose "Quick Action" and a build environment will open up.
Select "Workflow receives current FILES OR FOLDERS in FINDER".
From the long list of functions available drag "Get Selected Finder Items" over to the empty work space.
Then drag "Run Shell Script" under that. Choose "/bin/bash" and Pass Input "as arguments".
Then cut-and-paste the following script into the script window, and save the automator action as "Gcode_comments".
Start your cut-and-paste on the line below this one: #------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ # # GCode_comments shell script to be run by Mac OS X automator. Mark Harris Nov 2022. # #------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ empty_line="\n" degree="°" mm="mm" gcode_file=$1 pause="" filename_only=$(echo "$gcode_file" | sed s/"^.*\/"//) stl_filename=$(grep -m1 ";MESH" "$gcode_file" | grep -v "NONMESH" | sed s/"^.*[:]"//) machine_name=$(grep "Machine" "$gcode_file" | sed s/"^.*[ ]"//) nozzle=$(grep "Nozzle" "$gcode_file" | sed s/"^.*[ ]"//) material=$(grep "Material" "$gcode_file" | sed s/"^.*[ ]"//) temperature=$(grep "Temperature" "$gcode_file" | sed s/"^.*[ ]"//) slice_date=$(grep "Slice date" "$gcode_file" | sed s/"^.*[ ]"//) print_time=$(grep "Print time" "$gcode_file" | sed s/"^.*[ ]"//) adhesion=$(grep "Adhesion" "$gcode_file" | sed s/"^.*[ ]"// | sed s/"none"/NoBrim/) pause=$(grep "current layer" "$gcode_file" | sed s/"^.*[:]"// | sed s/" "/Pause/) if [ "$pause" = "" ]; then pause=$(grep "M600" "$gcode_file" | sed s/"^.*[n]"/Pause/) fi osascript -e "display alert \"$filename_only\" message \" Mesh: $stl_filename $empty_line $empty_line $nozzle$mm $material $temperature$degree $adhesion $pause $empty_line $empty_line $machine_name $empty_line Sliced $slice_date Print time $print_time \"" #------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ End your cut-and-paste on the line above this one.
The white space between the brackets in the lines that contain the function 'sed' should be explicit tab characters. I don't know if they will survive going through your browser and cut-and-paste.
If you have trouble, you can delete that white space between the brackets and hit your tab key between them.
The 'pause' lines detect the Cura extensions used to pause the printer for filament change, so the pop-up will also show if your G-code includes those.
Even if you're not a programmer, you can probably see the pattern in the code, and add or remove slicer conditions to suit your own needs.
And that's it ! You should now be able to right-click on your G-code files, and choose the quick-action called "gcode_comments". A pop-up Alert window should then open with selected slicer conditions displayed.
I'm sure Windows has a similar alert system, and if anyone wants to tell me how to set that up, I'd be happy to include their instructions here.
Note that cura does write out some conditions in the SETTINGS_3 records of the gcode file, but some are missing, and I just found it easier to parse my own records than theirs, in case you're wondering.
If you liked Gcode_comments, and you take photographs, you might also like MacMetaMod.
Mark Harris
Visits :